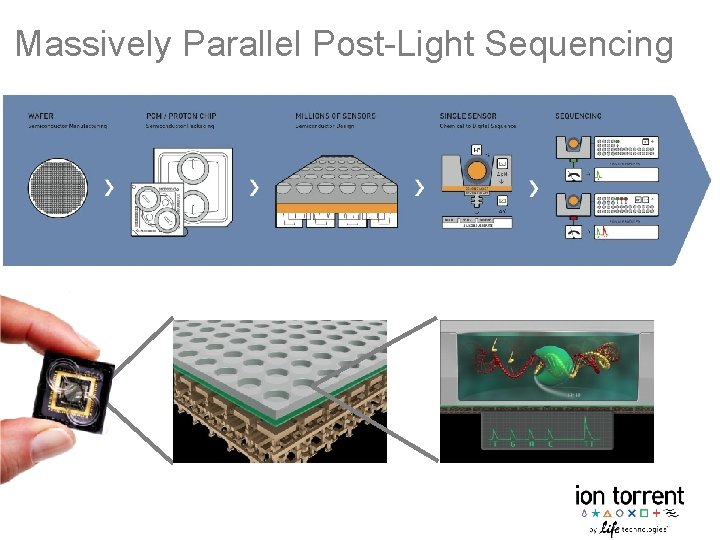
Ion semiconductor sequencing, a relatively recent advancement in DNA sequencing technology, has revolutionized genomics research and personalized medicine. This method, often referred to as ion torrent sequencing, relies on detecting the release of hydrogen ions (H+) during DNA synthesis rather than using fluorescent labels. This article will provide a comprehensive overview of the technology, focusing on its underlying principles, the patent landscape, and practical implications.
Fundamentals of Ion Semiconductor Sequencing
At its core, DNA sequencing involves determining the precise order of nucleotides (adenine, guanine, cytosine, and thymine - A, G, C, and T) within a DNA molecule. Traditional Sanger sequencing, while accurate, is relatively slow and expensive, limiting its scalability. Ion semiconductor sequencing offers a faster and more cost-effective alternative.
The Process Explained
The process can be broken down into several key steps:
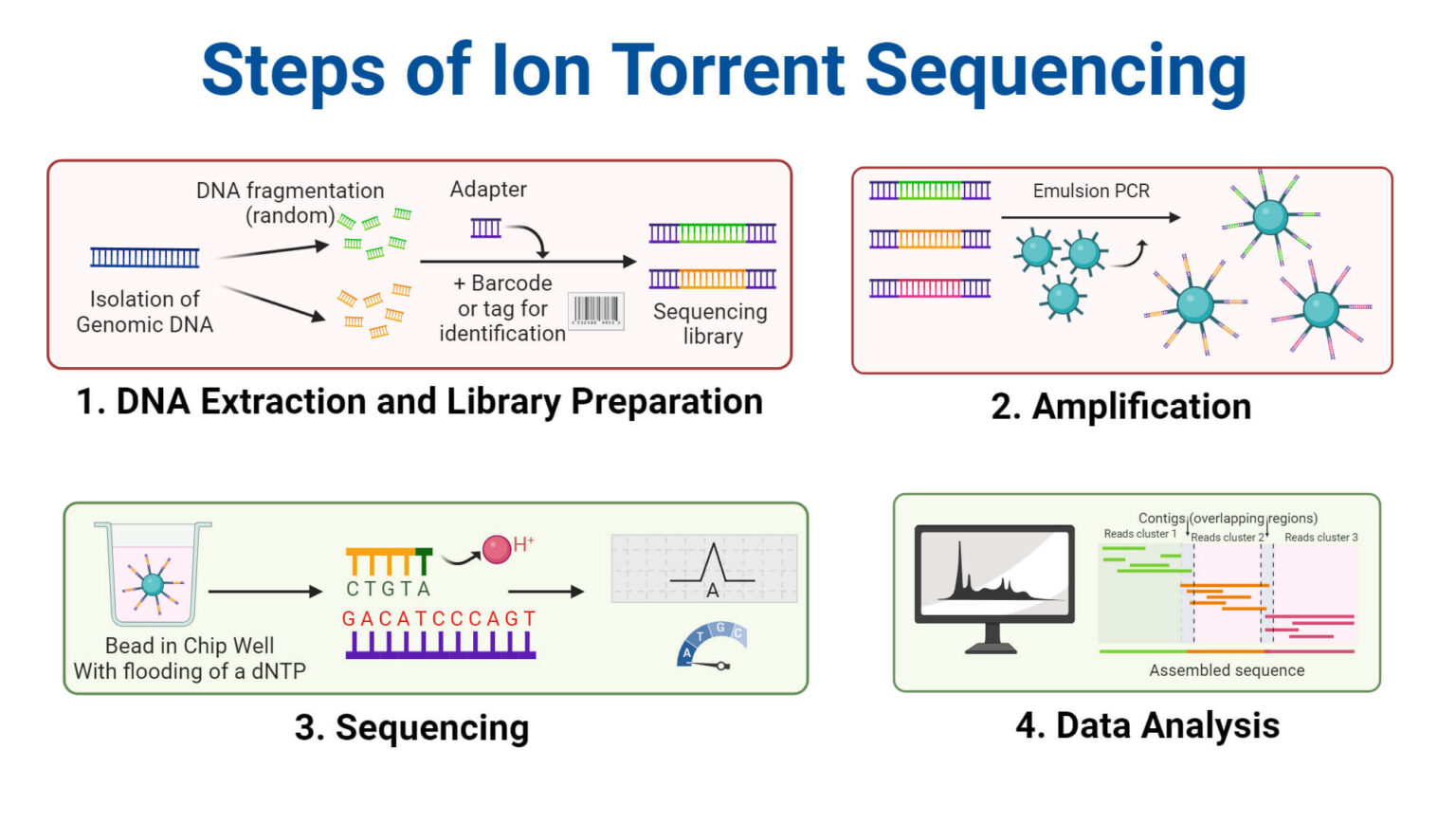
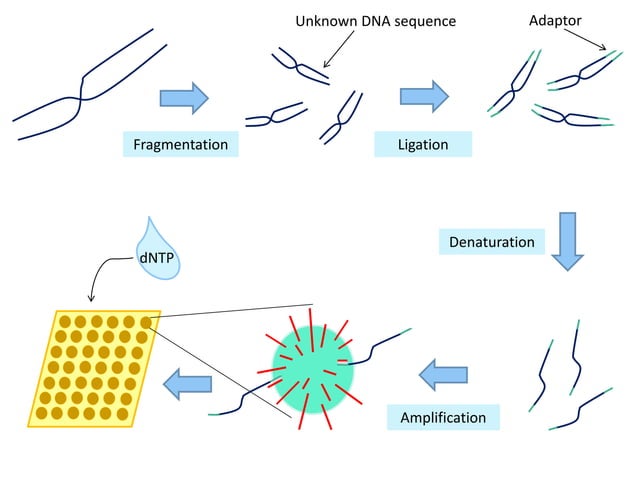
- DNA Fragmentation: The target DNA molecule is first fragmented into smaller, manageable pieces, typically a few hundred base pairs in length. This fragmentation is achieved through enzymatic or mechanical methods.
- Library Preparation: These fragments undergo library preparation, a process that involves attaching adapter sequences to the ends of the DNA fragments. These adapters are short, synthetic DNA sequences that serve as anchors for subsequent amplification and sequencing steps. They allow the DNA fragments to bind to the surface of the sequencing chip and also incorporate primer binding sites for PCR amplification.
- Emulsion PCR (emPCR): The adapter-ligated DNA fragments are then amplified using emulsion PCR. In emPCR, each DNA fragment is encapsulated within a microscopic aqueous droplet containing PCR reagents (DNA polymerase, nucleotides, primers) and a single bead. The bead is coated with primers complementary to the adapter sequences. PCR amplification occurs within each droplet, resulting in millions of copies of the DNA fragment attached to the bead. This creates clonal bead populations, each containing identical copies of a single DNA fragment.
- Enrichment: After emPCR, the beads containing amplified DNA fragments are enriched, meaning beads without DNA or with failed amplification are removed. This step ensures that the subsequent sequencing reaction is efficient and accurate.
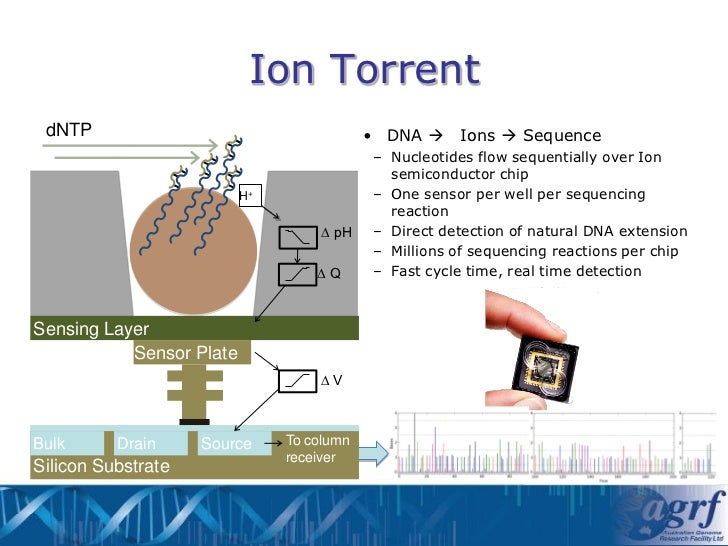
- Chip Loading: The enriched beads are then loaded onto a semiconductor sequencing chip. This chip contains millions of microwells, each capable of holding a single bead. Below each microwell is an ion-sensitive sensor.
- Sequencing by Ion Detection: Sequencing occurs through sequential flooding of the chip with each of the four nucleotide types (A, G, C, T). When a nucleotide complementary to the template strand is incorporated by DNA polymerase, a hydrogen ion (H+) is released as a byproduct. The ion sensor beneath the microwell detects this change in pH. The magnitude of the pH change is proportional to the number of nucleotides incorporated in a single flow cycle. For example, if the template strand contains a string of three adenines (AAA) and the chip is flooded with thymine (T), three thymines will be incorporated, releasing three hydrogen ions and generating a stronger signal.
- Data Analysis: The signals from the ion sensors are then processed by sophisticated algorithms to determine the sequence of the DNA fragment within each microwell. The sequence data is then assembled to reconstruct the original DNA sequence.
Advantages of Ion Semiconductor Sequencing
Compared to other sequencing technologies, ion semiconductor sequencing offers several advantages:
- Speed: It is generally faster than Sanger sequencing and many other next-generation sequencing (NGS) methods.
- Cost-Effectiveness: The absence of expensive fluorescent labels and optical detection systems reduces the cost per base.
- Scalability: The technology is easily scalable, allowing for the sequencing of entire genomes or targeted regions of interest.
- Real-Time Monitoring: The sequencing process can be monitored in real-time, allowing for immediate data analysis.
Patent Landscape of Ion Semiconductor Sequencing
The development and commercialization of ion semiconductor sequencing are protected by a complex web of patents. These patents cover various aspects of the technology, including the sequencing chips, the methods for library preparation, and the algorithms for data analysis.
Key Patent Areas: The key areas of patent protection relate to:
- The design and fabrication of the semiconductor sequencing chips, specifically the ion-sensitive sensors and the microfluidic systems for nucleotide delivery.
- The methods for preparing DNA libraries, including the adapter sequences, the emPCR protocols, and the enrichment strategies.
- The algorithms and software used to analyze the data generated by the sequencing chips, including base calling, error correction, and sequence alignment.
Example Patent Application Attributes:
A representative patent application will typically disclose:
A detailed description of the semiconductor chip architecture, including the materials used, the dimensions of the microwells, and the design of the ion-sensitive sensors.
The methods for manufacturing the chip, including the lithographic techniques used to create the microstructures.
The protocols for preparing DNA libraries, including the adapter sequences, the emPCR conditions, and the enrichment procedures.
The algorithms used to process the data generated by the sequencing chips, including the base calling algorithms, the error correction algorithms, and the sequence alignment algorithms.
Experimental data demonstrating the performance of the technology, including the accuracy, the speed, and the throughput of the sequencing system.
Practical Applications and Everyday Insights
Ion semiconductor sequencing has numerous applications in various fields, including:
- Medical Diagnostics: Identifying genetic mutations associated with diseases, such as cancer and inherited disorders. For example, rapid sequencing of tumor DNA can guide treatment decisions by identifying actionable mutations.
- Personalized Medicine: Tailoring medical treatments to an individual's genetic profile. This includes selecting the most effective drugs and dosages based on a patient's genetic makeup.
- Infectious Disease Surveillance: Monitoring the spread of infectious diseases by rapidly sequencing the genomes of pathogens. This helps track outbreaks, identify drug-resistant strains, and develop effective interventions.
- Agricultural Biotechnology: Improving crop yields and disease resistance by sequencing the genomes of plants and animals. This can lead to the development of more sustainable and productive agricultural practices.
- Forensic Science: Identifying individuals based on their DNA profiles. This is used in criminal investigations, paternity testing, and other forensic applications.
Insights for Everyday Life: While most people won't directly interact with ion semiconductor sequencing technology, its impact is increasingly felt in various aspects of life. Consider the following:
- Understanding Health Information: With the increasing availability of genetic testing services, understanding the basics of DNA sequencing can help individuals interpret their own genetic information and make informed decisions about their health.
- Appreciating Medical Advancements: Appreciating the role of sequencing technologies in the development of new diagnostics and treatments for diseases like cancer can foster a greater understanding of medical progress.
- Supporting Scientific Research: Supporting research in genomics and related fields can contribute to the further development and application of these technologies, leading to improved health outcomes and a better understanding of the world around us.
- Consider Ethical Implications: Be mindful of the ethical considerations surrounding genetic information, such as privacy and potential discrimination. Informed discussions and policies are crucial to ensure responsible use of this powerful technology.
In conclusion, ion semiconductor sequencing represents a significant advancement in DNA sequencing technology, offering speed, cost-effectiveness, and scalability. Its wide range of applications and increasing accessibility are transforming various fields, from medicine to agriculture. As the technology continues to evolve, its impact on our lives will only grow stronger. A grasp of its fundamental principles, the associated patent landscape, and its practical applications empowers individuals to engage more effectively with the rapidly evolving world of genomics.